Application of Genomic Tools in Plant Breeding
Transform agriculture with cutting-edge genomic tools. Discover real-life case studies on CRISPR's role in improving crop traits. Explore today!

Modern agricultural science has revolutionized crop development through advanced genomic methodologies. Historically, enhancing plant varieties demanded extensive experimental cycles spanning multiple years. Contemporary genetic analysis techniques dramatically accelerate selective breeding processes. Researchers now utilize sophisticated molecular screening approaches to identify optimal genetic combinations with unprecedented precision. In this article, we'll explore the different types of tools available and their applications in crop improvement. We'll also cover 2 case studies to learn how genomic tools are transforming agriculture.
Types of Genomic Tools Used in Plant Breeding
Next-Generation Sequencing
Next-generation sequencing(NGS) is one of the most powerful tools in modern plant breeding. It allows breeders to sequence the entire genome of a plant, providing an in-depth look at its genetic makeup. This technology enables the identification of genes associated with important traits, such as disease resistance, drought tolerance, and improved nutritional content. With NGS, breeders can select parent plants that are more likely to pass on these desirable traits to their offspring. In addition, CRISPR sequencing services offer advanced gene-editing capabilities, enabling breeders to modify specific genes associated with beneficial traits. By combining NGS with CRISPR technology, breeders can accelerate the development of improved crops with enhanced characteristics, optimizing both breeding efficiency and crop productivity.
Marker-Assisted Selection (MAS)
Marker-assisted selection (MAS) represents an advanced genetic screening methodology for identifying plant varieties with precise genomic characteristics. Unlike traditional phenotypic assessments that evaluate observable physical attributes, MAS employs molecular genetic markers to detect inherent trait potentials, such as enhanced agricultural resilience or productivity potential.
This sophisticated genomic approach significantly accelerates selective breeding protocols by enabling researchers to rapidly identify and propagate genetically superior plant specimens. By circumventing extensive field evaluation processes, MAS minimizes temporal and resource investments while optimizing genetic improvement strategies.
Genome-wide association Studies
Genome-wide association Studies (GWAS) provide a robust computational methodology for identifying genomic variations correlated with specific phenotypic characteristics. By conducting comprehensive genomic analyses across substantial plant populations, researchers can precisely map genetic loci responsible for critical agricultural traits, including pathogen resistance, biomass accumulation, and reproductive yield potential.
CRISPR/Cas9 Technology
One of the most groundbreaking genomic tools is CRISPR, a gene-editing technology that allows scientists to make precise changes to an organism's DNA. In plant breeding, CRISPR can be used to enhance desirable traits like pest resistance, drought tolerance, and even nutritional value. At CD Genomics, we offer CRISPR mutation sequencing and off-target validation services to ensure the accuracy and precision of CRISPR-edited plants.
Phenotype analysis and high-throughput phenotyping techniques
High-throughput phenotyping technology (HTP), combined with modern equipment such as drone sensors, can generate large amounts of data about plant growth dynamics. Combined with genotype data, these data can provide a more comprehensive understanding of plant phenotypic characteristics and their relationship to genetic background.
Genomic tools are profoundly transforming the field of plant breeding, providing new possibilities for improving crop yield, quality and stress resistance. These technologies not only accelerate the application of traditional breeding methods but also provide strong support for solving global food security issues.
Applications in Plant Breeding
The application of genomic tools in plant breeding mainly focuses on the following aspects: improving yield and quality, enhancing disease resistance and tolerance, and improving nutritional value.
Improve the yield and quality
Genome tools can significantly improve crop yield and quality through precise gene editing technology. For example, gene editing technology can increase crop yields by modifying specific genes, while optimizing fruit quality, such as extending shelf life, improving flavor and color. In addition, genomic technology can also be used to develop new crop varieties that not only have higher yields, but also can meet consumers 'needs for high-quality food.
Enhance disease resistance and tolerance
Genomic tools play an important role in improving crop disease resistance and tolerance. For example, gene editing technologies such as CRISPR can accurately modify plant genomes, giving plants resistance to multiple pathogens such as viruses, bacteria, fungi, and nematodes. In addition, genomic technology can also help plants adapt to various environmental pressures, such as drought, salt-alkali, cold and heat stress. These characteristics make genomic tools an important means to deal with global climate change and sustainable agricultural development.
Improving nutritional value
In recent years, genomic tools have also made significant progress in crop nutrition fortification. For example, through gene editing technology, scientists can increase nutrients such as beta-carotene and vitamins A, C and E in crops, thereby improving the nutritional value of foods. In addition, genomic technology could also be used to develop crops rich in health-promoting compounds, such as those with low gluten content, which can help reduce the risk of allergies and improve food safety.
Case Studies of Genomic Tools in Plant Breeding
Case Study 1: Gene editing without foreign DNA opens up a new way for raspberry breeding
Background
Raspberry (Rubus idaeus) is a high-value horticultural crop whose global production has increased significantly in recent years. It has the characteristics of high heterozygosity, asexual reproduction, etc., and traditional breeding cycles are long and often limited. Protoplast-based gene editing is particularly critical for the asexual reproduction of crops because it is difficult for these crops to remove foreign fragments through routine genetic transformation followed by backcross isolation. In particular, crops with outcrossing and high heterozygosity such as raspberries, cannot maintain genetic consistency through seed reproduction. In order to achieve accurate gene editing while avoiding the transfer of foreign DNA, the "DNA-free" method for CRISPR/Cas9 editing is used in raspberries, which can retain the excellent genetic background of commercial lines and meet the requirements of non-GMO products. Regulatory needs.
Methods
Raspberry roots were treated at 4°C for 50+ days to enhance protoplast yield. Protoplasts were isolated from stem cultures and prepared by enzymatic digestion, vacuum infiltration, and shaking. Purification used W5 solution centrifugation and sucrose gradient separation. For transfection, protoplasts were incubated with Cas9, gRNA, lipofectamine, and PEG, then washed and resuspended for DNA extraction to sequencing after 24 hours.

Figure 1. Protocol for protoplast isolation in raspberry. (Ryan Creeth et.al ,2025)
Results
Analysis of Sanger results using TIDE showed that the editing efficiency obtained at the PDS1 target was about 14%, and the predicted indels ranged from +1 to -5. Although the T7EI for the PDS2 target was not significant, TIDE statistics still yielded an editing rate of about 6.2%. NGS results showed that after NGS was performed on the same batch of PCR products, the results showed that the PDS1 editing rate was approximately 19%, and different types of deletions were detected, ranging from -28 bp to +1 bp. The most common indels are 6% of the 3bp deletion, and there are some differences in where DSBs occur; the PDS2 editing rate is approximately 2.3%, and the mutation frequency is lower than TIDE estimates.

Figure 2. Estimated indels were predicted by Sanger sequencing/ TIDE deconvolution, and actual indels were detected by NGS. (Ryan Creeth et.al ,2025)
Case Study 2: Gene editing technology was used to create a new wheat strain with high yield and disease resistance
Background
As one of the major food crops, wheat feeds more than one-third of the world's population. Powdery mildew is one of the major diseases affecting wheat yield worldwide. Due to the pleiotropic nature of the gene, the MLO gene is not only a susceptible gene to powdery mildew but also affects other physiological characteristics of wheat. The researchers found that while wheat mlo mutant showed resistance to powdery mildew, it also showed negative phenotypes such as premature aging and reduced yield, which may limit its widespread use in production. So how to create wheat varieties that have both disease resistance and yield?
Results
When the research team screened a wheat mutant population with the MLO gene, they found a new MLO mutant strain, Tamlo-R32. While showing resistance to powdery mildew, its growth, development and yield were completely normal. The results of whole-genome sequencing showed that the Tamlo-R32 mutant strain had a large 304-kb base pair deletion at the second exon position of the TaMLO-B1 locus.
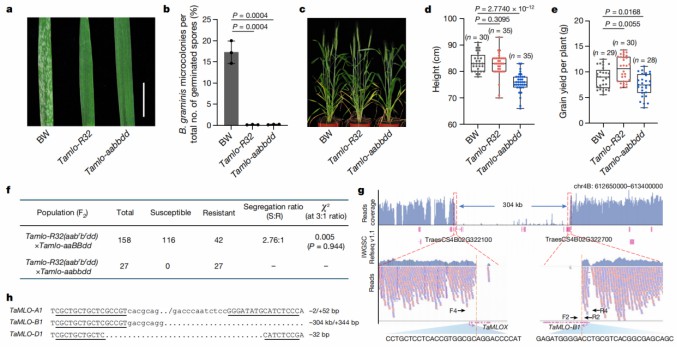
Figure 3. The Tamlo-R32 mutant strain showed resistance without affecting growth and yield. (Li, et al ,2022)
In order to study the impact of this large deletion, the research team used RNA-seq, qRT-PCR and other methods to detect the expression of genes near the deletion site. The sequencing results showed that the expression of related genes was down-regulated as expected or could not be detected. However, it is worth noting that the expression of the TaTMT3B gene upstream of the deletion site is significantly upregulated.
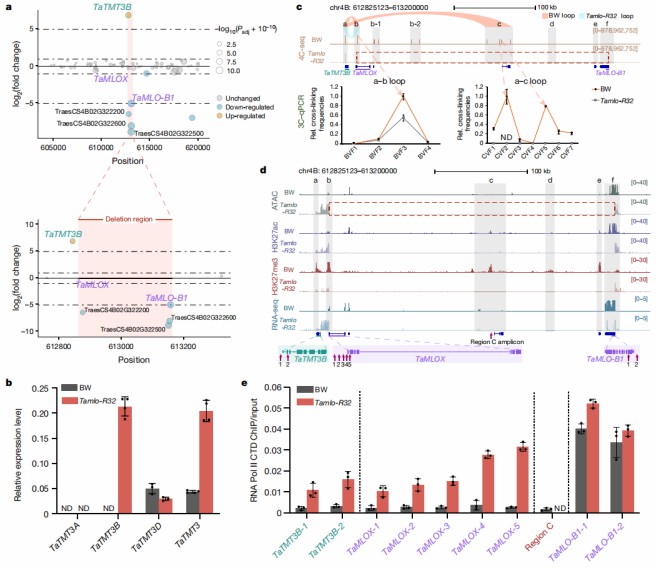
Figure 4. Altered chromatin status in Tamlo-R32 mutants leads to upregulation of the TaTMT3B gene (Li, S. et al ,2022)
After revealing the mechanism by which the Tamlo-R32 mutation can maintain yield, the research team tried to introduce the mutation into wheat. First, the Tamlo-R32 mutation was introduced into the main wheat varieties through traditional breeding methods, and it was found that it could significantly improve the powdery mildew resistance of these varieties. However, ordinary genetic transformation methods are time-consuming and labor-intensive. In order to complete this task more efficiently, the research team then used CRISPR-Cas9 technology to achieve targeted mutations and successfully obtained broad-spectrum powdery mildew resistance in just 2-3 months. Wheat varieties whose growth and yield are not affected confirm the application value of the Tamlo-R32 mutant gene in production.
What's Your Reaction?



















